Difference between revisions of "SNParrayDB"
(→Using SNParrayDB) |
(→Scan a genomic region for SNPs) |
||
| Line 6: | Line 6: | ||
===Scan a genomic region for SNPs=== | ===Scan a genomic region for SNPs=== | ||
[Section in progress] | [Section in progress] | ||
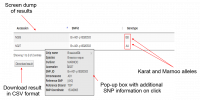
| − | [[File:Mypic]] | + | [[File:Mypic.png|200px|thumb|left|alt text]] |
===Bulk download SNPs by chromosome=== | ===Bulk download SNPs by chromosome=== | ||
Revision as of 07:57, 7 February 2018
Contents
Introduction
Advances in sequencing technology have led to a rapid rise of available genomic data for plants, driving new insights into the evolution and domestication of crops. Single nucleotide polymorphisms are a major component of crop diversity and are invaluable as genetic markers in breeding programs. To address the lack of SNP data provided by major databases and to make this data more easily accessible to researchers and breeders, we have developed SNParrayDB, the first database which focuses on SNP assay data from oilseed rape (Brassica napus) and bread wheat (Triticum aestivum). SNParrayDB is a comprehensive web-portal providing a database of large-scale genome variation from the Brassica 60K Illumina Infinium™ assay for 446 lines from 19 countries and from the Infinium iSelect Wheat 90k array for 309 lines. We include data from three recently published SNP assay datasets (Mason et al., 2015; Qian et al., 2014; Xu et al., 2016) and provide search, download and upload utilities for users. SNParrayDB is a useful starting point for a range of genomics and molecular breeding activities for oilseed rape and wheat.
Using SNParrayDB
Scan a genomic region for SNPs
[Section in progress]
Bulk download SNPs by chromosome
[Section in progress]
Links
The Crop SNParrayDB can be accessed here: http://snpdb.appliedbioinformatics.com.au:5000/
References
- Mason AS, Zhang J, Tollenaere R, Vasquez Teuber P, Dalton-Morgan J, Hu L, Yan G, Edwards D, Redden R, Batley J. 2015. High-throughput genotyping for species identification and diversity assessment in germplasm collections. Mol Ecol Resour 15(5): 1091-1101.
- Qian LW, Qian W, Snowdon RJ. 2014. Sub-genomic selection patterns as a signature of breeding in the allopolyploid Brassica napus genome. BMC Genomics 15.
- Xu LP, Hu KN, Zhang ZQ, Guan CY, Chen S, Hua W, Li JN, Wen J, Yi B, Shen JX, et al. 2016. Genome-wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.). DNA Research 23(1): 43-52.
- The Triticeae Toolbox (T3) (http://triticeaetoolbox.org/)
Back to main page